
Just in time for the weekend, we have lots of fantastic feedback via scientist ?alexh? from the Allen Institute for Brain Science. He has been providing everyone with frequent comments on each challenge to date. Have you been regularly checking in on your traces? (You may be surprised. He often points out areas that need tracing, and paying attention to those can boost your score.) Whenever possible, he also tries to provide us with a summary of “how we did” afterwards.
In case you missed it from the forums, the "gold standard" label is given to things that have already been currently reconstructed by experts (either elsewhere or at the Allen Institute for Brain Science) that we use to evaluate against to measure the effectiveness of our tools and help identify areas that could use more education efforts.
Mouse-V1-04
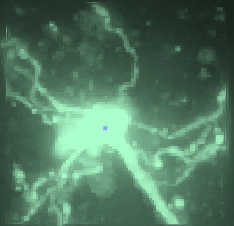 This is a spiny cell, with essentially no axon (the axon exits the soma and is immediately cutoff at the surface of the slice). The final consensus failed to capture only four short segments, but most importantly there were zero false positives.
This is a spiny cell, with essentially no axon (the axon exits the soma and is immediately cutoff at the surface of the slice). The final consensus failed to capture only four short segments, but most importantly there were zero false positives.
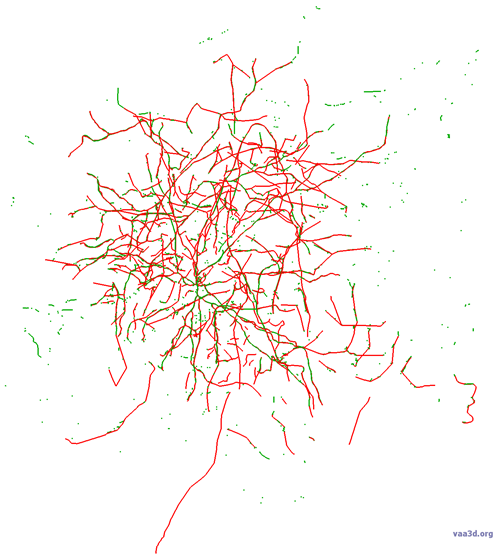
Below is an image of the overlay with consensus in green and gold standard in red.
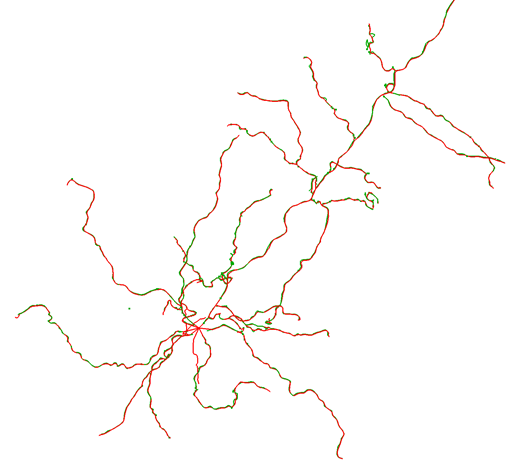
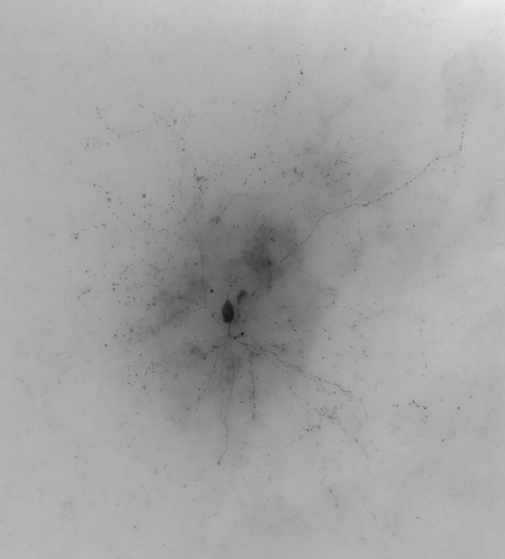
Because they agree so closely, you may find it difficult to see the consensus in some places. The reasons we think this consensus was so successful include an excellent signal-to-noise in the images (which can be seen in the maximum intensity projection (MIP) below-right), and no axon present.
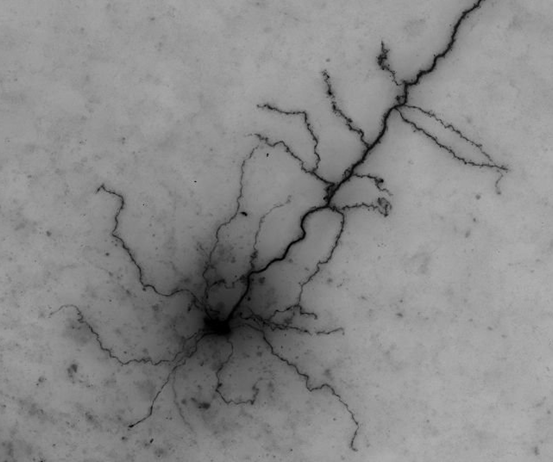
Mouse-V1-05
 This is a spiny cell, with a decent amount of axon. Our gold standard does not include the axon for this cell, but I can still do a good comparison. Only one dendritic segment was missed, and it was partially obscured by the soma, so it is an understandable miss. The axon captured in the consensus looks great. This consensus would be an excellent starting point for us. It would definitely save us time compared with the autotrace as a starting point. There are a few false positives (not enough to be problematic), including the bubble edge that we discussed last month. Here is the overlay image and the MIP (note that the axon is often invisible from the MIP image):
This is a spiny cell, with a decent amount of axon. Our gold standard does not include the axon for this cell, but I can still do a good comparison. Only one dendritic segment was missed, and it was partially obscured by the soma, so it is an understandable miss. The axon captured in the consensus looks great. This consensus would be an excellent starting point for us. It would definitely save us time compared with the autotrace as a starting point. There are a few false positives (not enough to be problematic), including the bubble edge that we discussed last month. Here is the overlay image and the MIP (note that the axon is often invisible from the MIP image):
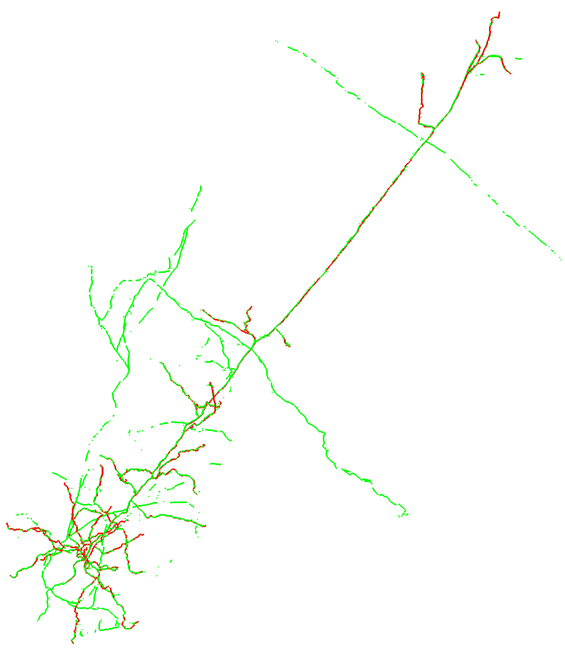
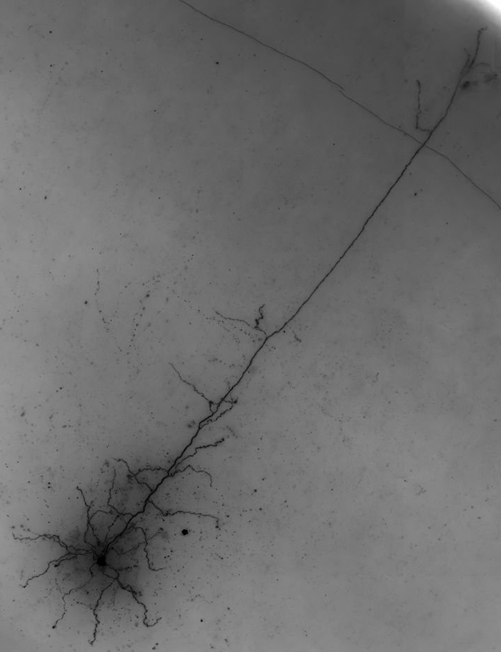
Mouse-V1-06
 This is an aspiny cell with tons of axon. Importantly, there is not much false positive in the consensus. The dendrites were mostly captured, though a few segments, branches and tips were missed. There was a lot of missed axon, with about 60% captured. Of note, there was a bunch of axon that was missed in the upper-left of the image. The reason for this is that there was no connection back to the main structure and the gap was larger than the fog would allow tracers to see. However, as is, the consensus would be a good starting point for us.
This is an aspiny cell with tons of axon. Importantly, there is not much false positive in the consensus. The dendrites were mostly captured, though a few segments, branches and tips were missed. There was a lot of missed axon, with about 60% captured. Of note, there was a bunch of axon that was missed in the upper-left of the image. The reason for this is that there was no connection back to the main structure and the gap was larger than the fog would allow tracers to see. However, as is, the consensus would be a good starting point for us.
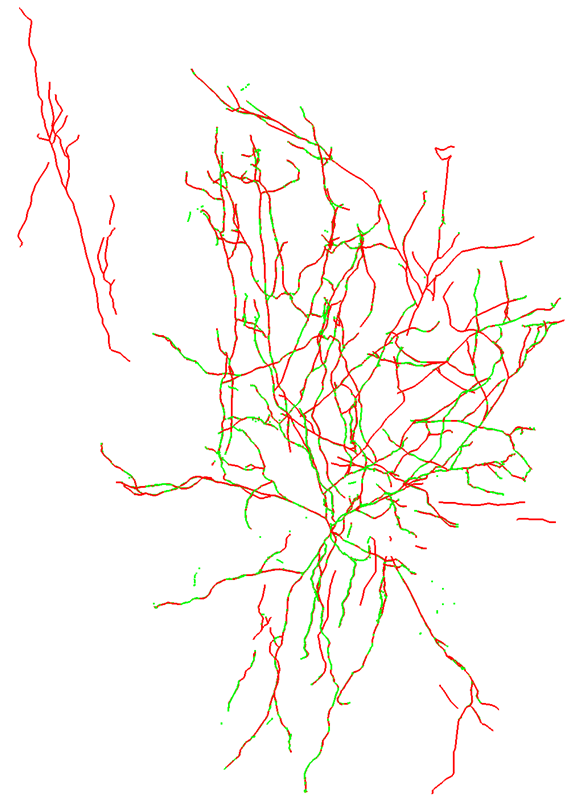
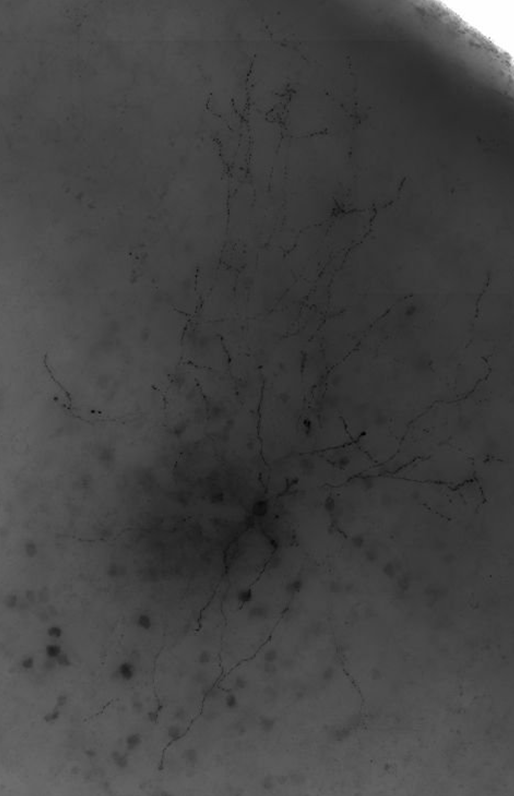
Mouse-V1-07
 This is an aspiny cell with poor image/cell quality and lots of axon. There is more missed dendrite than in a typical case (5 large segments). You can see from the maximum intensity projection (MIP) that even the dendrites are much more difficult to discern for this cell than previous cells. The axon is about 50% complete. There are numerous axon segments that are captured, but only as a series of dots (not a continuous line). There is also a lot of false positive signal throughout the consensus. We attribute this to the overall poor quality of the image/cell; low signal-to-noise is very difficult to reconstruct. This consensus is a great starting place.
This is an aspiny cell with poor image/cell quality and lots of axon. There is more missed dendrite than in a typical case (5 large segments). You can see from the maximum intensity projection (MIP) that even the dendrites are much more difficult to discern for this cell than previous cells. The axon is about 50% complete. There are numerous axon segments that are captured, but only as a series of dots (not a continuous line). There is also a lot of false positive signal throughout the consensus. We attribute this to the overall poor quality of the image/cell; low signal-to-noise is very difficult to reconstruct. This consensus is a great starting place.
Bring us your science questions on the General Discussion Forum.
Eager for feedback on earlier challenges? Scientist notes on 03: Mouse Neuron V1 is available for review.
Look for another feedback wrapup post on Monday. Good job, everyone, and thank you alexh for taking time out to send post-challenge notes and your continued feedback so we can keep learning.



